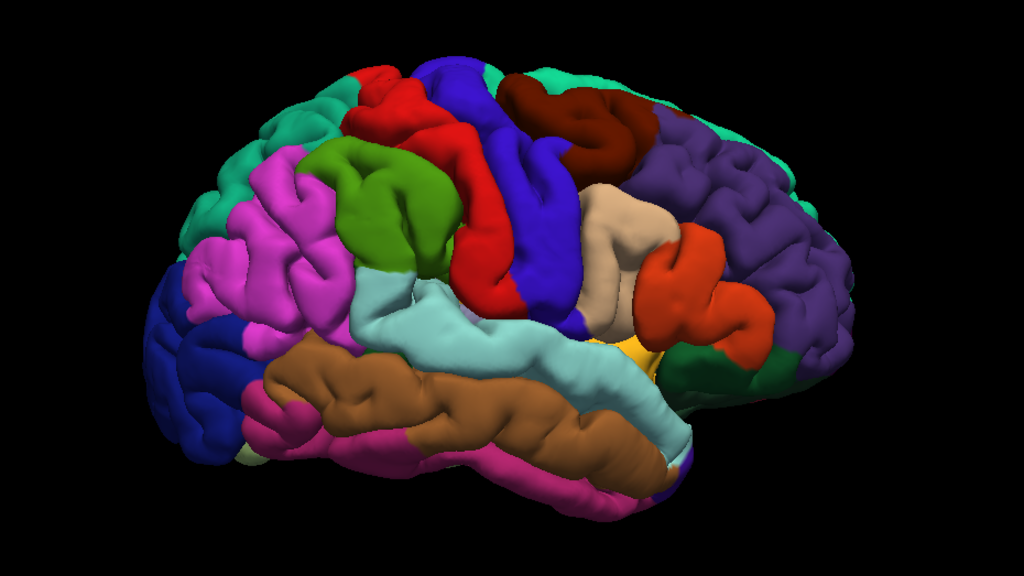
Anatomical Pipelines
INC Anatomical Pipeline
The INC standard anatomical processing pipeline, includes:
- Reorientation
- Denoising
- Rigid alignment to common space
- Coregistration to base anatomical (usually T1w)
- Within-modality averaging
- FG/BG parsing
- Intensity bias correction
- Brain extraction
- Intensity scaling
- Normalization to standard space(s)
Cost is determined by the number of anatomical contrasts (e.g., T1w, T2w, FLAIRw, etc.).
BRAINSAutoWorkup
The BRAINSAutoWorkup pipeline optimizes tissue classification through an iterative framework and produces robust parcellation of brain regions including in multi-site settings.
Labeling procedure
- Multi-atlas
- Similarly weighted
- Majority-vote (joint label fusion)
- Expert-segmented templates (adapted from Desikan-Killiany)
Brain regions included
- Cotrical and subcortical regions
- Where appropriate, separated by:
- Hemispheres
- Tissue type (gray or white matter)
FreeSurfer
FreeSurfer is a set of software tools for the study of cortical and subcortical anatomy.
In the cortical surface stream, the tools construct models of:
- The boundary between white matter and cortical gray matter
- The pial surface
Once these surfaces are known, an array of anatomical measures becomes possible, including:
- Cortical thickness
- Surface area
Voxelwise Maps
WM Anomalies
Estimated using Sequence Adaptive Multimodal SEGmentation (SAMSEG) implemented with FreeSurfer, they provide:
- Robustly segmentation of dozens of brain structures without preprocessing
- A map of anomalous WM regions
- WM voxels where the intensity is brighter (FLAIR) or darker (T2*) than cortical gray matter are marked as anomalous
- Segmentation of GM, WM, and large subcortical structures
They require:
- Work best with T1w images and either FLAIR- or T2*-weighted images
T2* and T1rho
Both T2* and T1rho image processing provide:
- Estimated values by weighting and combining multiple acquisitions varying in echo time (TE)
They involve:
- Coregistering to each participant/session native space
- Normalizing to template space
Jacobian Determinants
Maps of Jacobian Determinants of deformation matrices provide:
- A measurement of the magnitude of the difference in local, voxelwise volume for each participant/session
- A standard neurological template
- Distortion correction in voxel-based morphometry
- Variables in tensor-based morphometry
They require:
- Completed normalization to the desired template
Myelin Content
Myelin maps are calculated as a standardized ratio of T1w and T2w intensities. They provide:
- An estimate related to the local myelin content in each voxel
They require:
- Coregistered T1w and T2w images
- Normalization to a standard space for group analyses
Anatomical Labeling
Lesion Mapping
Manual identification and labelling of gross anatomical lesions involves:
- A first pass through SAMSEG
- Manual inspection and alteration of lesion boundaries
- Consensus among two independent lesion mappers for final lesion maps
Tissue Segmentation
Classification of tissue into N categories based on signal intensity provides:
- A three-category classification and maximum likelihood labels for CSF, GM, and WM
- Posterior probabilities for each class
Cerebellar Lobules
Lobule parcellation involves:
- Registering anatomical images using a pre-registered exemplars in a combined, registration quality metric
- Normalizing to a set of labelled exemplars from the COBRALab [1]
- A Joint Label Fusion approach
- Propagating labels to individual participant native space using a weighted majority vote
- Weights correspond to local intensity similarity between the normalized brain and exemplars
Thalamic Subregions
Parcellation of subnuclei of the thalamus, as implemented in Freesurfer, provides:
- A parcellation of the thalamus into 25 different nuclei, using a probabilistic atlas built with histological data
It requires a structural MRI basis as either:
- The main T1 scan processed through recon-all
- An additional scan of a different modality, which potentially shows better contrast between the nuclei
Iglesias et al., 2018 [2]
Medial Temporal Lobe
Parcellation of hippocampal subfields and amygdalar nuclei provides:
- An automated segmentation of the hippocampal substructures [3] and the nuclei of the amygdala [4]
It requires:
- Implementation in FreeSurfer
- A probabilistic atlas [5] built with ultra-high resolution ex vivo MRI data (~0.1 mm isotropic)
Brainstem
Parcellation of brainstem substructures provides:
- An automated segmentation of four different brainstem structures
- Medulla oblongata, pons, midbrain and superior cerebellar peduncle (SCP)
It requires:
- Implementation in FreeSurfer
- A Bayesian segmentation algorithm [6] that:
- Relies on a probabilistic atlas of the brainstem (and neighboring brain structures)
- Is built upon manual delineations of the structures on interest in 49 scans
- 10 for the brainstem structures
- 39 for the surrounding structures
References
[1] Min Tae M. Park, Jon Pipitone, Lawrence H. Baer, Julie L. Winterburn, Yashvi Shah, Sofia Chavez, Mark M. Schira, Nancy J. Lobaugh, Jason P. Lerch, Aristotle N. Voineskos, M. Mallar Chakravarty, Derivation of high-resolution MRI atlases of the human cerebellum at 3 T and segmentation using multiple automatically generated templates, NeuroImage, Volume 95, 15 July 2014, Pages 217-231, ISSN 1053-8119.
[2] Iglesias JE, Insausti R, Lerma-Usabiaga G, Bocchetta M, Van Leemput K, Greve DN, Van der Kouwe A, Fischl B, Caballero-Gaudes C, Paz-Alonso PM, Alzheimer's Disease Neuroimaging Initiative. A probabilistic atlas of the human thalamic nuclei combining ex vivo MRI and histology. Neuroimage. 2018 Dec 1;183:314-26.
[3] A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: Application to adaptive segmentation of in vivo MRI. (1) Iglesias, J.E., Augustinack, J.C., Nguyen, K., Player, C.M., Player, A., Wright, M., Roy, N., Frosch, M.P., Mc Kee, A.C., Wald, L.L., Fischl, B., and Van Leemput, K. Neuroimage, 115, July 2015, 117-137.
[4] High-resolution magnetic resonance imaging reveals nuclei of the human amygdala: manual segmentation to automatic atlas. Saygin ZM & Kliemann D (joint 1st authors), Iglesias JE, van der Kouwe AJW, Boyd E, Reuter M, Stevens A, Van Leemput K, Mc Kee A, Frosch MP, Fischl B, Augustinack JC. Neuroimage, 155, July 2017, 370-382.
[5] Bayesian longitudinal segmentation of hippocampal substructures in brain MRI using subject-specific atlases. Iglesias JE, Van Leemput K, Augustinack J, Insausti R, Fischl B, Reuter M. Neuroimage, 141, November 2016, 542-555.
[6] Bayesian segmentation of brainstem structures in MRI. Iglesias, J.E., Van Leemput, K., Bhatt, P., Casillas, C., Dutt, S., Schuff, N., Truran-Sacrey, D., Boxer, A., and Fischl, B. NeuroImage, 113, June 2015, 184-195.